
- Labview 2013 and windows 10 software#
- Labview 2013 and windows 10 code#
- Labview 2013 and windows 10 Offline#
Labview 2013 and windows 10 code#
The LabVIEW code can be compiled into an executable file if wished. It must be noted that the user needs a licensed copy of the basic LabVIEW software. While LabVIEW contains the same concepts found in most traditional programming languages, such as different data types, loops, variables, and object-oriented programming, the visual representation allows for easy access and modification of the code, in contrast to programming languages where the code is written in text. LabVIEW is a graphical programming language, where code is written by wiring together graphical modules.
Labview 2013 and windows 10 Offline#
INMEX can perform much more advanced meta-analysis, ADE allows working offline with large datasets, easy modification of the code (see below), and to prioritize and exclude array spots according to their specificity. Here, we present a LabVIEW program, Array Data Extractor (ADE), which allows users to extract expression information for a list of genes from multiple datasets, merge it into one output file, and perform basic statistics.

A remarkable online tool, INMEX, has recently been published, providing user-friendly web-based platform for meta-analysis, but other available tools require substantial bioinformatics skills perform cross-platform meta-analysis.
Labview 2013 and windows 10 software#
Whilst other software packages allow very advanced data processing, performing a meta-analysis with data from multiple studies and platforms is still difficult for a “bench” scientist, and there is a lack of user-friendly software allowing researchers do so in a fast and easy way. Other examples of open source software are the TM4 Microarray Software Suite and GenePattern. the open source software Bioconductor, allowing for complex microarray analysis like pre-processing, quality assessment, differential expression, clustering and classification, and gene set enrichment analysis. Various approaches have been published to normalize and refine data to detect meaningful expression changes in genes/networks and there are several software packages, e.g. Combining and comparing data from different studies is a rewarding approach, but comparing data across several studies is a challenging task. This provides researchers with the opportunity to detect novel treatment targets for various diseases, discover and refine signaling pathways, and to identify unknown interaction networks. Today, there are close to one million preprocessed datasets publicly available repositories like the NCBI Gene Expression Omnibus, ArrayExpress or the Stanford Microarray Database. High-throughput gene expression array technologies are commonly used in biomedical research and provide huge amounts of data.
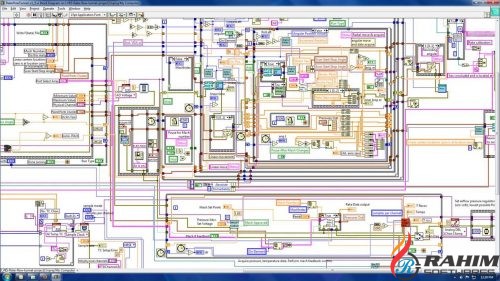
Further, the graphical programming language used in LabVIEW allows applying changes to the program without the need of advanced programming knowledge. ConclusionsĪlthough existing software allows for complex data analyses, the LabVIEW based program presented here, “Array Data Extractor (ADE)”, provides users with a tool to retrieve meaningful information from multiple normalized gene expression datasets in a fast and easy way.


Results confirmed known regulation of a beta 1 adrenergic receptor and further indicate novel research targets. Functionality was tested for 288 class A G protein-coupled receptors (GPCRs) and expression data from 12 studies comparing normal and diseased human hearts. Here, we present a user-friendly LabVIEW program to automatically extract gene expression data for a list of genes from multiple normalized microarray datasets. Highly advanced bioinformatics tools have been made available to researchers, but a demand for user-friendly software allowing researchers to quickly extract expression information for multiple genes from multiple studies persists. Large data sets from gene expression array studies are publicly available offering information highly valuable for research across many disciplines ranging from fundamental to clinical research.


 0 kommentar(er)
0 kommentar(er)
